Past Projects and Research
I spend a lot of my time learning and creating, and I’m trying to be better about documenting the process. To that end, this post will be a summary of some projects I worked on with colleagues while I was in school and when I first began to teach.
Project 1: Activity Recognition
Using physiological data collected by wearable technology, such as the acceleration and heat flux of a person’s body, this project intelligently characterizes the activity a user performed.
Wearable sensor technology has been investigated as a way to regularly monitor a person’s health from watches that track your heart rate during exercise, to wearable devices that use accelerometer data to check if someone has fallen or not. In this project, we use data, collected by wearable devices, to classify a person’s activity into one of three classes: sleeping, watching TV, and all other activities.
This follows the guidelines of the Physiological Data Modeling Contest (PDMC) contest, and you can read more about this data and the challenge in the project paper.
To approach this challenge, we compared unsupervised learning algorithms with supervised approaches. Our hypothesis was: a supervised algorithm would produce the most accurate test results, but a good, unsupervised algorithm would be ideal since it is time-intensive to create richly-labeled data for training.
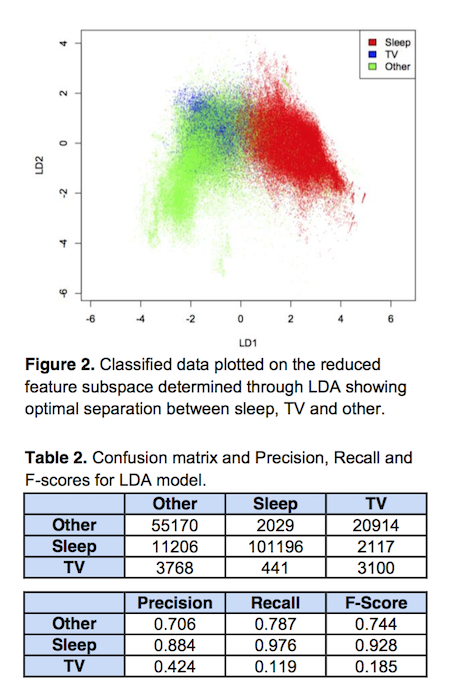
Linear Discriminant Analysis
In the image above, you can see an example of data separation done with linear discriminant analysis (LDA), which is a supervised learning model. The original data features included several different types of collected, sensor data; acceleration, heat flux, body temperature, and so on. In this case, LDA was trained on some training data and used as a feature reduction technique, which squashed the original features into 2D space while trying to maintain the greatest possible separation between classes. We decomposed the data into two new LDA components (linear combinations of the original features), and could then visualize these class groupings in space. You can see the separation between sleeping and other activity classes, but watching TV gets obscured.
We did get the best performance using supervised algorithms, but this project did get me thinking about how unsupervised approaches might be improved.
Project 2: Cell Detection
This project uses supervised learning algorithms and graph-mining techniques to automatically count and detect embryonic cells as they grow.
Cell detection and counting in the context of biology research is largely a labor-intensive, manual task. A reliable count is of paramount importance as it is often a fundamental step in various growth studies; most commonly in embryonic or cancer cell growth analysis. In this project, we studied the efficacy of SVM and graph-mining algorithms to automatically detect and count embryonic stem cells in fluorescent microscopy images. You can read about the detailed results, in this paper.
Graph Construction
To perform graph-mining, we constructed a graph that separated individual cells based on patterns of intensity in a given image. A cell nucleus was very bright, but its edges and the background were darker. So, in this approach, each cell in an image is modeled as a smooth 2D function that has a single local maximum in its neighborhood. We used this model to perform histogram segmentation.
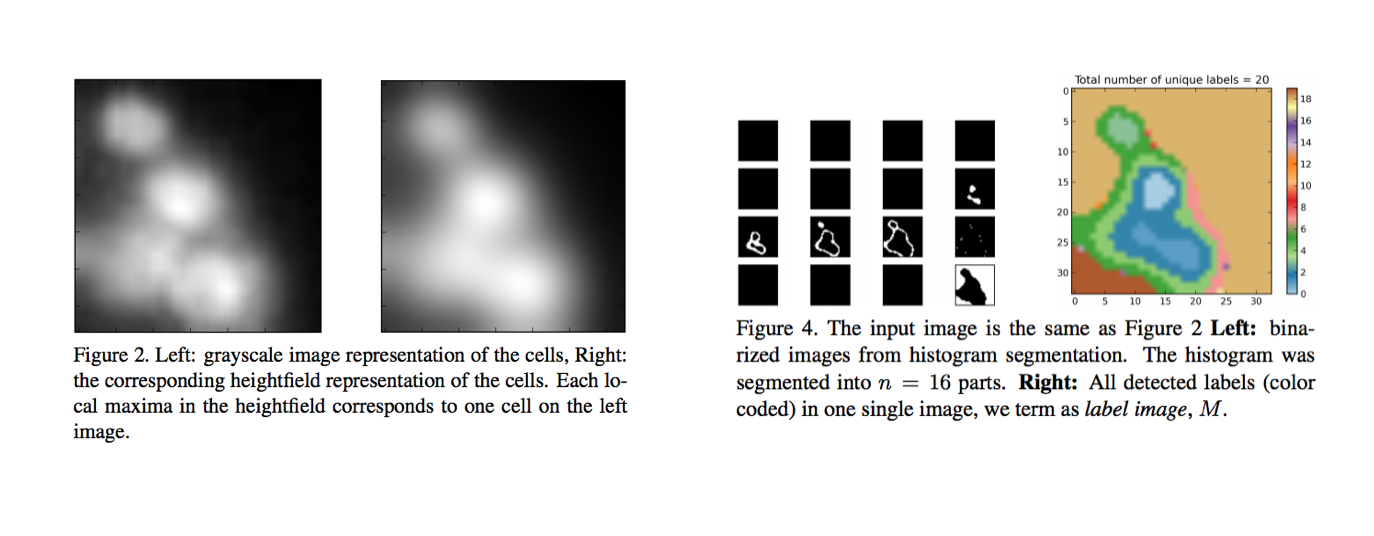
We traversed the histogram from the brightest range to the darkest, and assigned a numerical label to each pixel in the image, such that pixels with higher intensities have smaller label values (as you can see in the above image). Then we used these numbered segments to construct a graph.
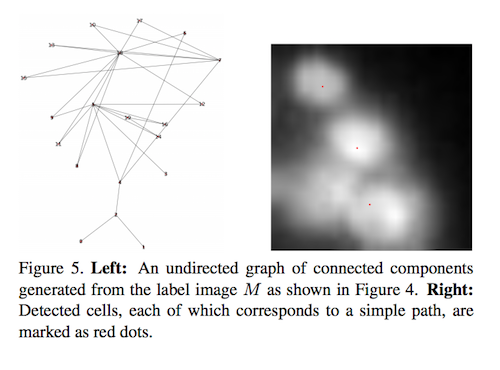
We used a matrix representation for this graph. Then, for each pixel in the segmented image, looked at four neighboring pixel labels to populate the matrix. Once the undirected graph was created, every cell in the input image had an associated simple path in that graph, and the intensity-peak of each cell was marked as the location of that cell. We also took some extra processing steps to account for multiple peaks in one cell, and to account for some cell overlap, and eventually found that graph-mining worked well.
Project 3: Depth Estimation
This project uses pairs of photos taken by an endoscope at slightly different depths and a SIFT algorithm to estimate the depth map for an area of interest.
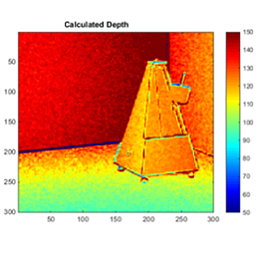
SIFT was used to create feature descriptors for two endoscopic images, taken at different depths. Using the SIFT descriptors, we identified the best-matching points in the pair of images and used those to calculate a transformation between the two images. From this transformation, we extracted an estimation for the distance between the end of the endoscope and the objects in the images.
Project 4: DNA-Based Logic Gates
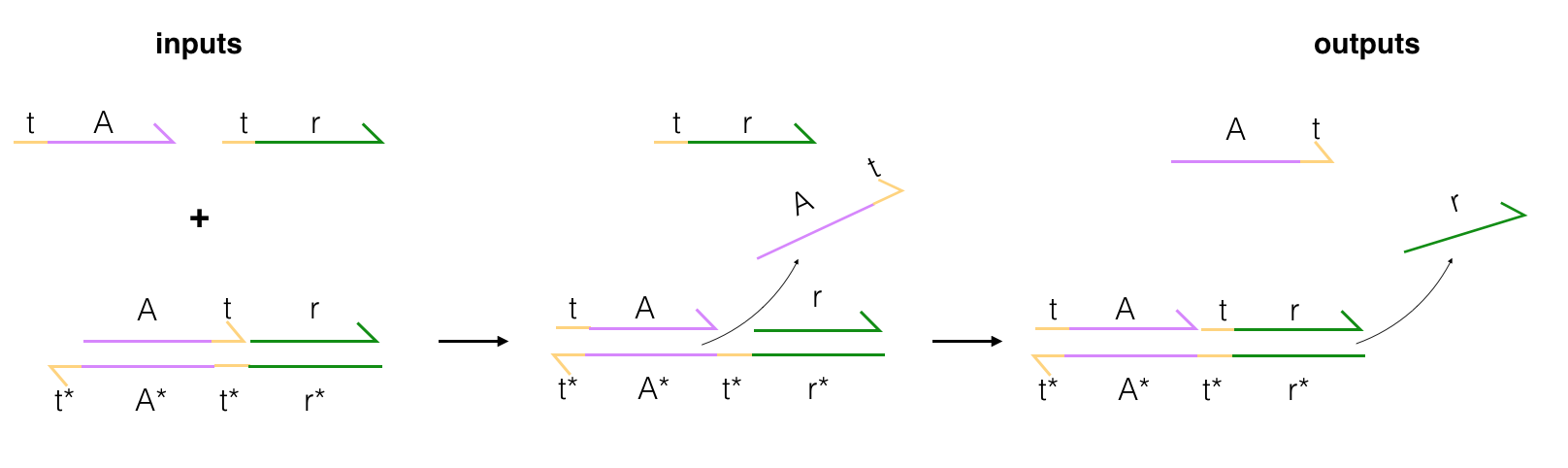
In my research as an undergraduate, I studied the construction of and uses cases for DNA-based logic gates. These gates rely on the predictable base-pairing of DNA to operate (A-T, C-G) and emulate the behavior of traditional logic gates: producing an output signal, given an input. The flow of a simple gate is pictured below, where A and A* are complementary strands of DNA. The gate wants to reach an equilibrium state in which all strands are perfectly paired with their match (ex. t-A is paired with t*-A*), and to reach this state, some displacement occurs.

Input strands are combined with a gate, and then these inputs start base-pairing with the gate; displacing the output strands. The final output strand typically has a fluorescent tag on it that can be measured once the strand is released (the signal is suppressed when the output strand is in a double-stranded configuration). Building off of this structure, pairs of gates can be used together to create AND and OR logic gates. These gates have the potential to be used in transistors or even in medicine that can target cells by their DNA/RNA markers.
Project 5: DNA-Mediated Lithography
I recorded a short, Vimeo video on lithography done in part by DNA-mediated etching of SiO2.
If you are interested in my work and thoughts, check out my Twitter or connect with me on LinkedIn.